Visualizing unstructured data
Example visualization of unstructured ICON and UGRID data.
Here we show, how the psy-maps plugin can visualize the unstructured grid of the Earth System Model ICON from the Max-Planck-Institute for Meteorology in Hamburg, Germany, and data following the unstructured grids (UGRID). The visualization works the same as for normal rectilinear grids. Internally, however, the coordinates are interpreted in a completely different way (see below for a detailed explanation).
[1]:
import psyplot.project as psy
import matplotlib as mpl
%matplotlib inline
%config InlineBackend.close_figures = False
[2]:
psy.rcParams['plotter.maps.xgrid'] = False
psy.rcParams['plotter.maps.ygrid'] = False
mpl.rcParams['figure.figsize'] = [10., 8.]
Visualizing UGRID data
A widely accepted approach for unstructured grids are the so-called UGRID Conventions. For a demonstration, we use the 'ugrid_demo.nc' netCDF file that contains sea surface height of a Tsunami simulation. We use the load parameter, to directly load it into memory. That speeds up the plotting of the data.
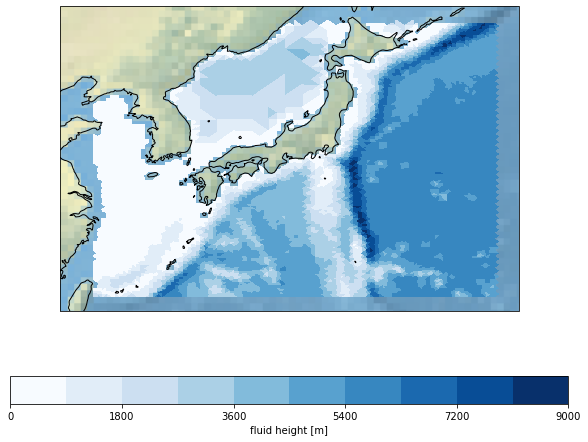
psyplot automatically recognizes the UGRID conventions and adapts it’s plotting algorithm for displaying the data. For this simulation, let’s focus on Japan. We use the maskleq keyword here to mask the land surface and display a stock_img on the continents.
[3]:
tsunami = psy.plot.mapplot(
'ugrid_demo.nc', name='Mesh2_height', load=True,
maskleq=0, lonlatbox='Japan', cmap='Blues',
clabel='{desc}', stock_img=True, lsm='50m')
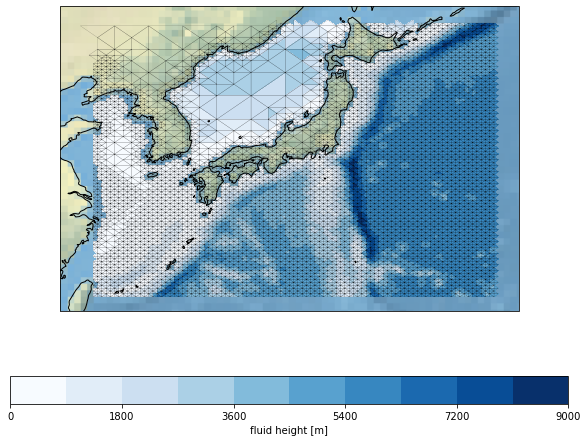
To visualize the unstructured grid, we can use the datagrid formatoption. It excepts a string or the line properties.
[4]:
tsunami.docs('datagrid')
datagrid
========
Show the grid of the data
This formatoption shows the grid of the data (without labels)
Possible types
--------------
None
Don't show the data grid
str
A linestyle in the form ``'k-'``, where ``'k'`` is the color and
``'-'`` the linestyle.
dict
any keyword arguments that are passed to the plotting function (
:func:`matplotlib.pyplot.triplot` for unstructured grids and
:func:`matplotlib.pyplot.hlines` for rectilinear grids)
See Also
--------
mask_datagrid: To display cells with NaN
See Also
--------
xgrid
ygrid
[5]:
tsunami.update(datagrid={'c': 'k', 'lw': 0.1})
tsunami.show()
[6]:
tsunami.close()
Visualizing scalar and vector ICON data
This section requires the psy-maps plugin and the file 'icon_grid_demo.nc' which contains one variable for the temperature, one for zonal and one for the meridional wind direction.
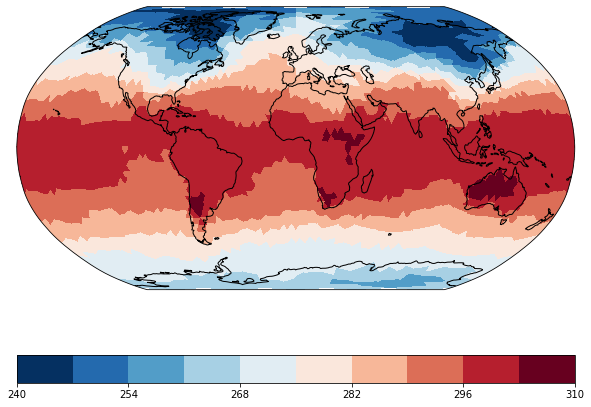
The visualization works the same way as for a usual rectangular grid. We choose a robinson projection and a colormap ranging from blue to red.
[7]:
maps = psy.plot.mapplot('icon_grid_demo.nc', name='t2m', projection='robin',
cmap='RdBu_r')
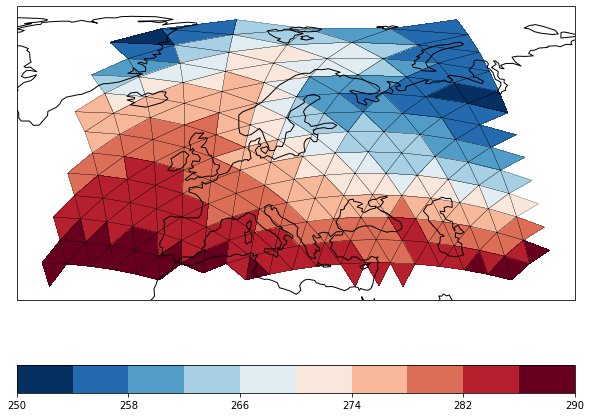
We can again zoom in to Europe and use the datagrid formatoption to display the triangular grid.
[8]:
maps.update(lonlatbox='Europe', datagrid=dict(color='k', linewidth=0.2))
maps.show()
The same works for vector data
[9]:
vectors = psy.plot.mapvector(
'icon_grid_demo.nc', name=[['u', 'v']] * 2, projection='robin',
ax=(1, 2), lonlatbox='Europe')
vectors.plotters[0].update(arrowsize=100)
vectors.plotters[1].update(plot='stream')
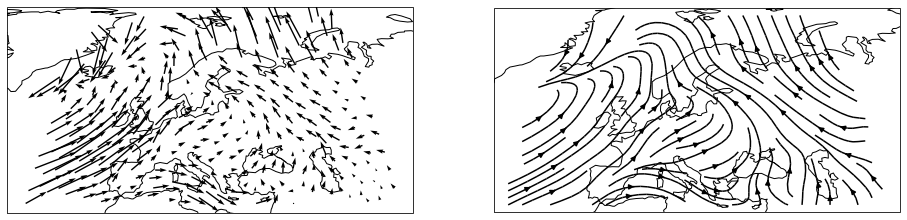
And combined scalar and vector fields
[10]:
combined = psy.plot.mapcombined(
'icon_grid_demo.nc', name=[['t2m', ['u', 'v']]], projection='robin',
lonlatbox='Europe', arrowsize=100, cmap='RdBu_r', datagrid={'c': 'k', 'lw': 0.1})
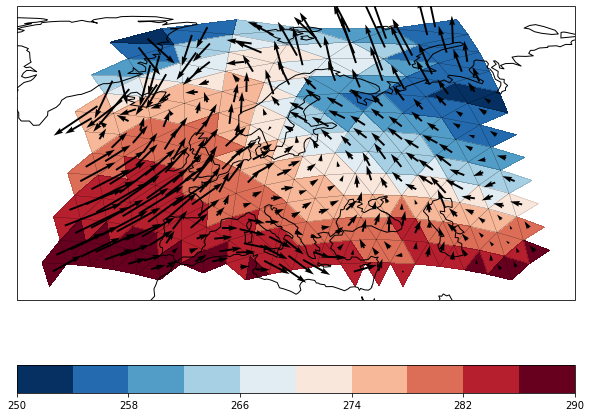
The mapplot plotmethod does also not care about the shape of the grid cells. Therefore it can also visualize the ICON edge grid:
[11]:
maps = psy.plot.mapplot('icon_grid_demo.nc', name='t2m_edge', projection='robin',
cmap='Wistia', datagrid=dict(c='b', lw=0.2))
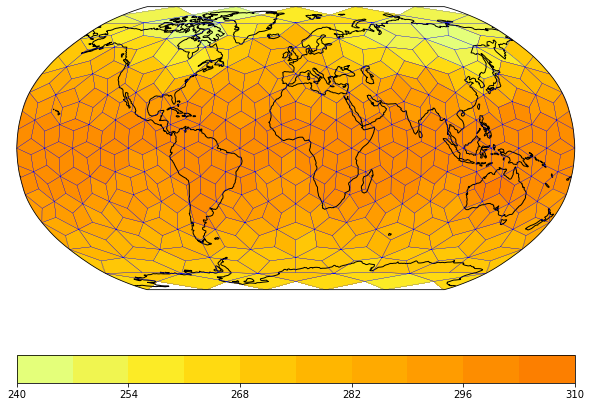
The handling unstructured grids
In this section, we go a bit into detail into how psyplot interpretes unstructured grids. If your data is an ICON file or follows the UGRID Conventions, you might want to skip this section, because then psyplot can handle your data automatically.
Interpreting the UGRID Conventions
The best way to specify unstructured grids is to follow the unstructured grids (UGRID) conventions. Variables that follow these conventions are then interpreted by the UGridDecoder class. If one variable has a mesh attribute, psyplot assumes that it follows the UGRID conventions.
Furthermore, psyplot interpretes the location attribute. Attribute can either live on the edge, face or the node of the grid cell. In case of a variable the lives on the node, the data is assumed to be triangular and we plot the data using a Delaunay triangulation.
[12]:
ds = tsunami[0].psy.base
ds
[12]:
<xarray.Dataset>
Dimensions: (nMesh2_node: 57222, nMesh2_face: 113885, Three: 3,
nMesh2_edge: 171106, Two: 2, time: 1)
Coordinates:
Mesh2 int32 -2147483647
Mesh2_node_x (nMesh2_node) float32 121.0 142.0 131.5 ... 131.5 136.8
Mesh2_node_y (nMesh2_node) float32 -59.2 -59.2 -52.03 ... -41.26 -44.85
Mesh2_face_nodes (nMesh2_face, Three) int32 0 1 2 2 1 ... 2 57219 57218 0 2
* time (time) datetime64[ns] 1950-01-01T04:11:59.999742507
* nMesh2_face (nMesh2_face) int64 0 1 2 3 ... 113882 113883 113884
Dimensions without coordinates: nMesh2_node, Three, nMesh2_edge, Two
Data variables:
Mesh2_edge_nodes (nMesh2_edge, Two) int32 1 2 2 0 0 ... 57218 2 57218 0
Mesh2_height (time, nMesh2_face) float32 4.069e+03 ... 4.117e+03
Mesh2_bathy (time, nMesh2_face) float32 6.931e+03 ... 6.883e+03
Mesh2_m_x (time, nMesh2_face) float32 -3.357e-10 ... -3.056e-10
Mesh2_m_y (time, nMesh2_face) float32 1.091e-10 ... -3.47e-10
Mesh2_u_x (time, nMesh2_face) float32 -8.25e-14 ... -7.422e-14
Mesh2_u_y (time, nMesh2_face) float32 2.681e-14 ... -8.429e-14
Mesh2_level (time, nMesh2_face) int16 7 8 9 10 10 11 ... 10 10 9 8 7
Attributes:
title: netCDF output from StormFlash2d
institution: University of Hamburg, KlimaCampus
contact: None
source: None
references: None
comment: None
Conventions: UGRID-0.9
creation_date: 2014-10-03 15:29:40 02:00
modification_date: 2014-10-03 15:29:40 02:00[13]:
print('Mesh variable of Mesh2_height:', ds.Mesh2_height.mesh)
print('Location of the variable in the grid cell:', ds.Mesh2_height.location)
print('Name of the location variable:', ds.Mesh2.face_node_connectivity)
print('--------------------------------------------')
print('face node connectivity:', ds.Mesh2_face_nodes)
Mesh variable of Mesh2_height: Mesh2
Location of the variable in the grid cell: face
Name of the location variable: Mesh2_face_nodes
--------------------------------------------
face node connectivity: <xarray.DataArray 'Mesh2_face_nodes' (nMesh2_face: 113885, Three: 3)>
array([[ 0, 1, 2],
[ 2, 1, 3],
[ 2, 3, 4],
...,
[57219, 2, 4],
[57218, 2, 57219],
[57218, 0, 2]], dtype=int32)
Coordinates:
Mesh2 int32 -2147483647
Mesh2_face_nodes (nMesh2_face, Three) int32 0 1 2 2 1 ... 2 57219 57218 0 2
* nMesh2_face (nMesh2_face) int64 0 1 2 3 ... 113882 113883 113884
Dimensions without coordinates: Three
Attributes:
cf_role: face_node_connectivity
long_name: Maps every triangular face to its three corner nodes
start_index: 0
Interpreting the CF Conventions
However, there is also another way that follows more closely the standard CF Conventions. This is also the way, that ICON uses, namely the netCDF attributes coordinates and bounds. These two attributes are decoded CFDecoder (namely the DataArray.psy.decoder attribute) and used for the visualization.
To explain it a bit more, we can look into the icon_grid_demo.nc file:
[14]:
ds = maps[0].psy.base
ds
[14]:
<xarray.Dataset>
Dimensions: (time: 5, ncells: 5120, vertices: 3, edge: 480, no: 4, lev: 4)
Coordinates:
* time (time) datetime64[ns] 1979-01-31T18:00:00 ... 1979-05-31T18:00:00
clon (ncells) float64 0.6283 0.5623 0.6283 ... 0.1454 0.2085 0.2409
clon_bnds (ncells, vertices) float64 0.6283 0.5671 0.6896 ... 0.2828 0.1899
clat (ncells) float64 0.9184 0.9399 0.8735 ... -0.7121 -0.7751 -0.7038
clat_bnds (ncells, vertices) float64 0.9626 0.8954 ... -0.6741 -0.6842
elon (edge) float32 0.6283 0.7726 0.484 ... 0.4396 0.4633 0.2847
elon_bnds (edge, no) float32 0.3907 0.6283 0.8659 ... 0.1728 0.2376 0.4088
elat (edge) float32 0.8232 0.9613 0.9613 ... -0.7843 -0.6409 -0.67
elat_bnds (edge, no) float32 0.809 0.7377 0.809 ... -0.6461 -0.809 -0.6887
* lev (lev) float64 1e+05 8.5e+04 5e+04 2e+04
* edge (edge) int64 0 1 2 3 4 5 6 7 ... 472 473 474 475 476 477 478 479
Dimensions without coordinates: ncells, vertices, no
Data variables:
t2m (time, lev, ncells) float32 ...
u (time, lev, ncells) float32 ...
v (time, lev, ncells) float32 ...
t2m_edge (time, lev, edge) float32 274.5 265.1 268.8 ... 212.1 212.4 212.1
Attributes:
CDI: Climate Data Interface version 1.9.1 (http://mpimet...
Conventions: CF-1.4
history: Thu Aug 30 21:54:23 2018: cdo delname,t2m_2d,u_2d,v...
number_of_grid_used: 42
uuidOfHGrid: bf575ad8-daa6-11e7-a4a9-93d511f821b4
title: Temperature and Wind demo-File for python nc2map mo...
CDO: Climate Data Operators version 1.9.1 (http://mpimet...This dataset contains two grid definitions, one for variables living on the face of one grid cell (namely t2m, u, v) and one for a variable living on the edges of a grid cell (t2m_edge). Which grid is chosen, used depends on the coordinates attribute of the specific variable:
[15]:
print('t2m:', ds.t2m.encoding['coordinates'])
print('t2m_edge:', ds.t2m_edge.encoding['coordinates'])
t2m: clat clon
t2m_edge: elat elon
The variables mentioned in these coordinates do then have a bounds attribute to the variable with the lat-lon information of the vortices for each grid cell
[16]:
print(ds.clat.bounds)
ds[ds.clat.bounds]
clat_bnds
[16]:
<xarray.DataArray 'clat_bnds' (ncells: 5120, vertices: 3)>
array([[ 0.962634, 0.895414, 0.895414],
[ 0.962634, 0.958974, 0.895414],
[ 0.895414, 0.828942, 0.895414],
...,
[-0.684167, -0.69024 , -0.755691],
[-0.755691, -0.818119, -0.746385],
[-0.746385, -0.674147, -0.684167]])
Coordinates:
clon (ncells) float64 0.6283 0.5623 0.6283 ... 0.1454 0.2085 0.2409
clon_bnds (ncells, vertices) float64 0.6283 0.5671 0.6896 ... 0.2828 0.1899
clat (ncells) float64 0.9184 0.9399 0.8735 ... -0.7121 -0.7751 -0.7038
clat_bnds (ncells, vertices) float64 0.9626 0.8954 ... -0.6741 -0.6842
Dimensions without coordinates: ncells, vertices[17]:
print(ds.elat.bounds)
ds[ds.elat.bounds]
elat_bnds
[17]:
<xarray.DataArray 'elat_bnds' (edge: 480, no: 4)>
array([[ 0.809014, 0.737659, 0.809014, 0.918438],
[ 0.809014, 0.99046 , 1.107149, 0.918438],
[ 1.107149, 0.99046 , 0.809014, 0.918438],
...,
[-0.809014, -0.87296 , -0.740435, -0.688687],
[-0.740435, -0.59862 , -0.530217, -0.688687],
[-0.530217, -0.646124, -0.809014, -0.688687]], dtype=float32)
Coordinates:
elon (edge) float32 0.6283 0.7726 0.484 ... 0.4396 0.4633 0.2847
elon_bnds (edge, no) float32 0.3907 0.6283 0.8659 ... 0.1728 0.2376 0.4088
elat (edge) float32 0.8232 0.9613 0.9613 ... -0.7843 -0.6409 -0.67
elat_bnds (edge, no) float32 0.809 0.7377 0.809 ... -0.6461 -0.809 -0.6887
* edge (edge) int64 0 1 2 3 4 5 6 7 ... 472 473 474 475 476 477 478 479
Dimensions without coordinates: noIdentification of unstructured variables
psyplot automatically detects, whether the variable is unstructured based on the above mentioned bounds coordinate. If the length of the second dimension (here vertices or no) is larger then 2, then it assumes an unstructured variable.
Alternatively, it assumes that the variable is unstructured if the optional CDI_grid_type netCDF attribute or the grid_type attribute is equal to 'unstructured':
[18]:
ds.t2m.CDI_grid_type
[18]:
'unstructured'
[19]:
psy.close('all')
Acknowledgement
Thanks @Try2Code for providing the ICON data and thanks to Stefan Vater and the Research Group for Numerical Methods in Geosciences from the University of Hamburg for providing the Tsunami simulation file.